Our laboratory focuses on developing novel single-molecule tools in single cells to study the structure, function and dynamics of macromolecular assemblies. For example, we developed single-molecule gene expression reporting systems and chromosomal DNA conformation markers to probe the dynamics of gene regulation and transcription in bacterial cells. We also pioneered the use of superresolution imaging to probe the bacterial cell division machinery. Recently we expanded our horizons by collaborating with experts of different fields in biology to map the spatial organization of the genome and epigenetic markers of single human cells, and to develop new single-molecule based technologies for sensitive early detection of cancer markers in blood samples. Four major research directions are detailed below.
Structure, function and dynamics of bacterial cell division complex
In bacteria, cell division is carried out by a highly conserved supramolecular complex, the divisome. Understanding the structure and function of the divisome is important for developing new antibiotics. Central to the divisome is a ring-like structure formed by the essential FtsZ protein, named the Z-ring. The Z-ring recruits all other divisome components, and possibly generates a mechanic force to constrict the inner membrane. However, it has been difficult to probe the in vivo structural organization of the Z-ring because of its small size and highly dynamic nature. We made our first major contribution to the field by illustrating the structural organization of the Z-ring in live E. coli cells using single-molecule based superresolution imaging. The structure provides an important foundation to understand how different components of the divisome are assembled and coordinated to carry out cell division. Next, we showed that there exists a multi-layered protein network residing in the cytoplasm, physically connecting the Z-ring to the chromosome to stabilize the Z-ring and coordinate cell division with chromosome segregation. Most importantly, we provide substantial evidence to demonstrate that, in contrary to the common belief, the Z-ring is not the major constrictive force generator.  Instead, septum closure rate is limited by cell wall growth, and further modulated by nucleoid segregation. Furthermore, we recently discovered that the Z-ring dynamically treadmills along the division plane. Treadmilling is the apparent movement of a polymer by the continuous polymerization at one end and depolymerization at the other end, with each individual monomer remaining stationery during the process. We found that Z-ring’s treadmilling dynamics is powered by FtsZ’s GTPase activity, and guides the spatiotemporal distribution of cell wall synthesis machineries so that a symmetric, smooth septum and consequently two hemisphere new cell poles could be built in a circular motion. Our work redefines the role of the Z-ring in bacterial cell division, and promotes a new, holistic view of the divisome.
Instead, septum closure rate is limited by cell wall growth, and further modulated by nucleoid segregation. Furthermore, we recently discovered that the Z-ring dynamically treadmills along the division plane. Treadmilling is the apparent movement of a polymer by the continuous polymerization at one end and depolymerization at the other end, with each individual monomer remaining stationery during the process. We found that Z-ring’s treadmilling dynamics is powered by FtsZ’s GTPase activity, and guides the spatiotemporal distribution of cell wall synthesis machineries so that a symmetric, smooth septum and consequently two hemisphere new cell poles could be built in a circular motion. Our work redefines the role of the Z-ring in bacterial cell division, and promotes a new, holistic view of the divisome.
- Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science. 2017 Feb 17;355(6326):744-747.
- Coltharp C, Buss J, Plumer TM, Xiao J. Defining the rate-limiting processes of bacterial cytokinesis. Proc Natl Acad Sci U S A. 2016 Feb 23;113(8):E1044-53.
- Buss J, Coltharp C, Shtengel G, Yang X, Hess H, Xiao J. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 2015 Apr;11(4):e1005128.
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS One. 2010 Sep 13;5(9):e12682.
Stochastic gene expression
Gene expression is inherently stochastic. How cells battle noise in gene expression in order to achieve robust growth and development has been the subject of intense studies. In the past decade, majority of studies focused on the intrinsic noise of gene expression, which are fluctuations caused by the intrinsic properties of biochemical reactions in gene expression at the low-copy number regime. To find out how cells repress intr insic noise using autoregulatory transcription factors, we developed a single-molecule gene expression reporter to monitor the stochastic expression of a transcription factor and its autoregulatory actions. We discovered that intrinsic noise has negligible effect on the overall gene expression noise. Instead, extrinsic noise, or fluctuations caused by different compositions of cellular factors in different cells, dominates noise in gene expression. Consequently, cells use negative autoregulation to counteract extrinsic noise in order to achieve a homogenous gene expression level. We recently further developed a new strategy to monitor simultaneously the correlative expression dynamics of two transcription factors in live cells, both at the single molecule level. This ability has not been possible before. The new, dual single-molecule reporter system allowed her to uncover two new cell fate potentials that were hidden in a classic bistable switch, which is a common building block of gene regulatory networks, and traditionally is viewed as only capable of directing two mutually exclusive cell fates. We experimentally mapped the corresponding potential landscape and switching kinetics between four different gene expression states, offering first-of-its-kind insight of how cells could encode multiple cell fates even with a limited circuitry to allow a high degree of adaptation and differentiation. These new findings significantly advance our understanding of the operational principle of gene regulatory networks.
insic noise using autoregulatory transcription factors, we developed a single-molecule gene expression reporter to monitor the stochastic expression of a transcription factor and its autoregulatory actions. We discovered that intrinsic noise has negligible effect on the overall gene expression noise. Instead, extrinsic noise, or fluctuations caused by different compositions of cellular factors in different cells, dominates noise in gene expression. Consequently, cells use negative autoregulation to counteract extrinsic noise in order to achieve a homogenous gene expression level. We recently further developed a new strategy to monitor simultaneously the correlative expression dynamics of two transcription factors in live cells, both at the single molecule level. This ability has not been possible before. The new, dual single-molecule reporter system allowed her to uncover two new cell fate potentials that were hidden in a classic bistable switch, which is a common building block of gene regulatory networks, and traditionally is viewed as only capable of directing two mutually exclusive cell fates. We experimentally mapped the corresponding potential landscape and switching kinetics between four different gene expression states, offering first-of-its-kind insight of how cells could encode multiple cell fates even with a limited circuitry to allow a high degree of adaptation and differentiation. These new findings significantly advance our understanding of the operational principle of gene regulatory networks.
- Fang X, Liu Q, Bohrer C, Hensel Z, Han W, Wang J, Xiao J. Cell fate potentials and switching kinetics uncovered in a classic bistable genetic switch. Nat Commun. 2018 Jul 17;9(1):2787
- Feng H, Hensel Z, Xiao J, Wang J. Analytical calculation of protein production distributions in models of clustered protein expression. Phys Rev E Stat Nonlin Soft Matter Phys. 2012 Mar;85(3 Pt 1):031904.
- Hensel Z, Feng H, Han B, Hatem C, Wang J, Xiao J. Stochastic expression dynamics of a transcription factor revealed by single-molecule noise analysis. Nat Struct Mol Biol. 2012 Aug;19(8):797-802.
Spatial organization of genome and transcriptome
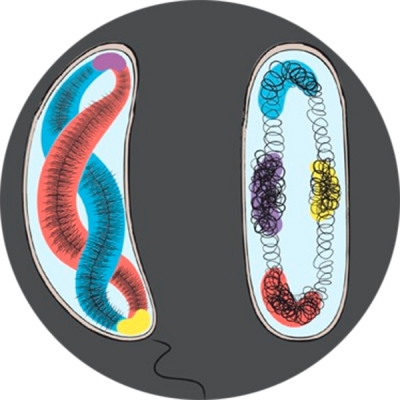
Recent studies suggest that genes are spatially organized; where they are and how they are organized are important for their transcription activities. To test this hypothesis, we are developing new imaging techniques to visualize chromosomal DNA conformation and transcription activity in individual cells. The first set of DNA localization markers we developed allowed us to probe the dynamics of transcription factor-mediated DNA looping in live E. coli cells, and relate to transcription activity of the gene it controls. We are currently developing new superresolution methods to probe both genome organization and transcription activity in both bacterial and eukaryotic stem cells. We expect this line of work to provide new insight into the spatial regulation of transcription.
- Weng X, Bohrer CH, Bettridge K, Lagda AC, Cagliero C, Jin DJ, Xiao J. Spatial organization of RNA polymerase and its relationship with transcription in Escherichia coli. Proc Natl Acad Sci U S A. 2019 Oct 1;116(40):20115-20123. doi: 10.1073/pnas.1903968116. Epub 2019 Sep 16. PubMed PMID: 31527272; PubMed Central PMCID: PMC6778201.
- Weng X, Xiao J. Spatial organization of transcription in bacterial cells. Trends Genet. 2014 Jul;30(7):287-97. doi: 10.1016/j.tig.2014.04.008. Epub 2014 May 23.
- Hensel Z, Weng X, Lagda AC, Xiao J. Transcription-factor-mediated DNA looping probed by high-resolution, single-molecule imaging in live E. coli cells. PLoS Biol. 2013;11(6):e1001591.
Developing cool imaging and analysis tools
Single-molecule localization based superresolution imaging has opened a wide door for biologists to explore the structures and dynamics of cellular  assemblies at unprecedented resolutions. We develop computational methods and quantitative imaging analyses to counter blinking behaviors of fluorescent labels, correct confinement errors, and identify spatial features of cellular structures. Furthermore, we are developing new single-molecule imaging based assays to visualize the activity, localization and identity of protein factors and chromosomal DNA sequences.
assemblies at unprecedented resolutions. We develop computational methods and quantitative imaging analyses to counter blinking behaviors of fluorescent labels, correct confinement errors, and identify spatial features of cellular structures. Furthermore, we are developing new single-molecule imaging based assays to visualize the activity, localization and identity of protein factors and chromosomal DNA sequences.
- Bohrer CH, Yang X, Weng X, Tenner B, Ross B, McQuillen R, Zhang J, Roberts E, Xiao J. A Pairwise Distance Distribution Correction (DDC) algorithm for blinking-free super-resolution microscopy. BioRxiv. 2019 September; doi: https://doi.org/10.1101/768051.
- Mo GC, Ross B, Hertel F, Manna P, Yang X, Greenwald E, Booth C, Plummer AM, Tenner B, Chen Z, Wang Y, Kennedy EJ, Cole PA, Fleming KG, Palmer A, Jimenez R, Xiao J, Dedecker P, Zhang J. Genetically encoded biosensors for visualizing live-cell biochemical activity at super-resolution. Nat Methods. 2017 Apr;14(4):427-434.
- Bohrer CH, Bettridge K, Xiao J. Reduction of Confinement Error in Single-Molecule Tracking in Live Bacterial Cells Using SPICER. Biophys J. 2017 Feb 28;112(4):568-574.
- Xiao J, Dufrêne YF. Optical and force nanoscopy in microbiology. Nat Microbiol. 2016 Oct 26;1(11):16186. doi: 10.1038/nmicrobiol.2016.186.
